Gallery
This page is still under development.
Research Projects
Postdoctoral Fellow (2022 - 2023)
University of Manitoba
I led the epidemiological analyses of projects in Nairobi, Kenya, aimed at enhancing our understanding of the epidemiology and transmission dynamics of HPV among female sex workers and men who have sex with men (MSM). I also used bioinformatics workflows, with a focus on developing pipelines for bulk and single-cell RNA sequencing, and trained PhD students on the workflows. In addition, I developed interactive Shiny projects and automated dashboards to improve monitoring and facilitate effective communication.
SWOP HPV Study
I contributed to the implementation of a longitudinal study among female sex workers and MSM clients at a SWOP clinic in Nairobi. Our main objective is to investigate the epidemiological aspects and mucosal/immunological characteristics of HPV genotypes, specifically examining their association with clearance over a one-year follow-up period. By delving into these factors, we anticipate that our findings will not only enhance the control of HPV and associated cancer burdens but also potentially contribute to the development of therapeutic vaccines.
In an earlier paper, we found a alarmingly high rates of HIV and HPV prevalence in a Kenyan MSM population and preliminary analysis from this study also suggests similar findings, calling for targeted HPV vaccination in both populations.
I also developed a dashboard for Prof. Keith Fowke’s Lab to monitor their newly launched IIQ2-Phase2 study in Kenya.
Xpert Magic Shiny App
I have developed Xpert Magic, an interactive app developed for the SWOP HPV Study project, where GeneXpert machines are used to determine HPV infection status. It is a powerful tool designed to effortlessly convert raw outputs of HPV and CTNG results from GeneXpert machines into tidy tabular data. Users can effortlessly upload their raw outputs to the app, which automatically processes the data and generates a formatted spreadsheet for downstream analyses. Experience the convenience of Xpert Magic, developed to streamline downstream data workflow.
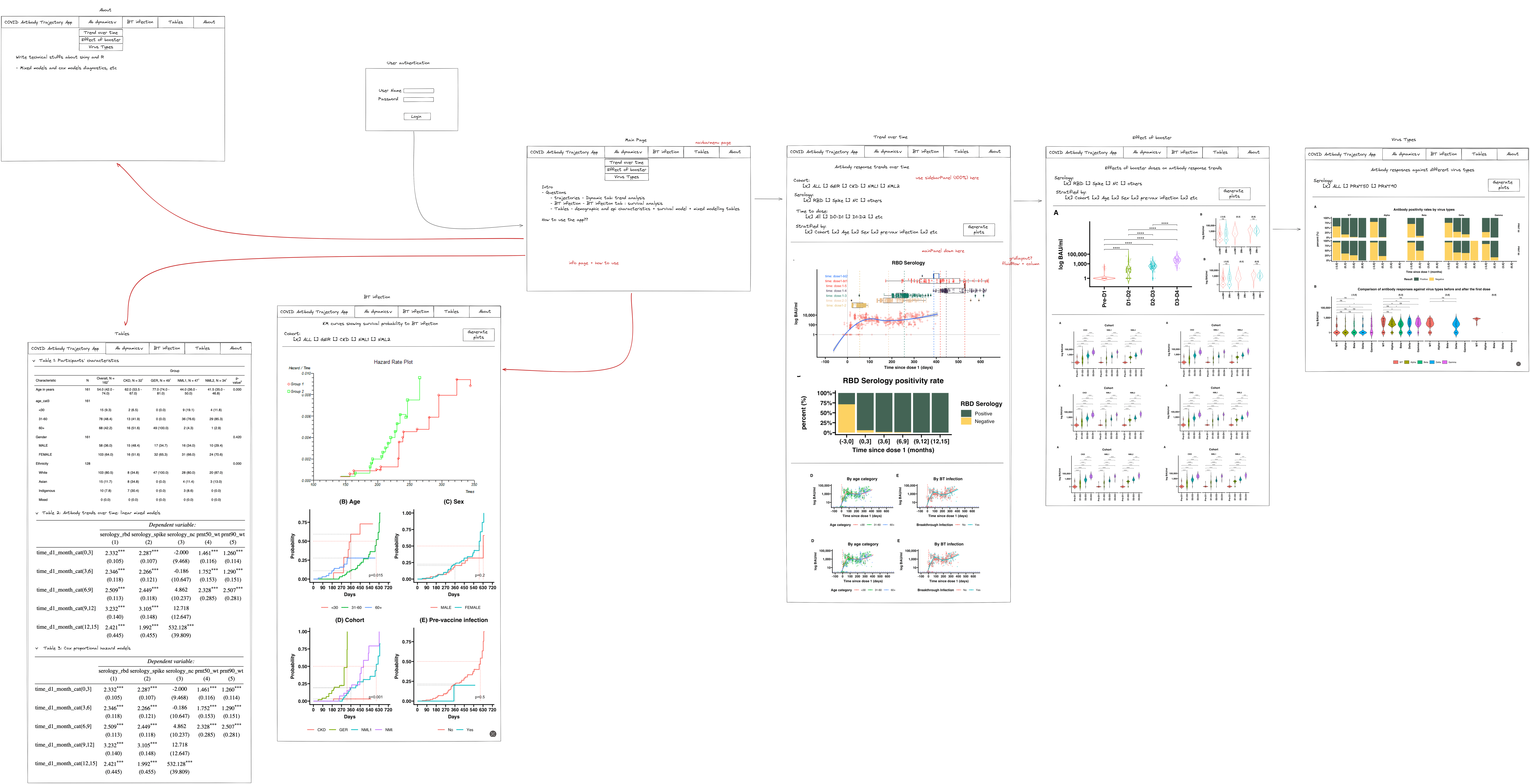
COVID-19 Antibody Trajectories
I led the epidemiological analysis of SARS-CoV-2 antibody trajectories post-vaccination, specifically examining the patterns and trends up to 3-5 booster doses. This research aims to shed light on the long-term effectiveness and potential benefits of multiple booster doses in enhancing immune responses against the virus. By understanding these trajectories, we can inform public health strategies and optimize vaccination protocols to better protect individuals and control the spread of COVID-19.
In addition, I have developed a Shiny app as a communication tool to facilitate cohort analysis and showcase the antibody trajectories of our research. This app serves as a comprehensive platform to present the outcomes of our study, which include three types of serology antibodies and various neutralization responses. Given the complexity and multiple dimensions of these outcomes, generating statistics and graphs for each one can be challenging. However, the Shiny app streamlines the process, providing interactive visualizations and data exploration capabilities for a more engaging and accessible presentation of our findings. This app allows us to effectively communicate and collaborate with our research partners, enabling a deeper understanding of the antibody trajectories and their implications for immune response against SARS-CoV-2. It truly represents the cherry on top of our contributions, enhancing our ability to share and disseminate our research outcomes.
Colp Study
This study investigates the interplay of tissue resident memory T cells and their compositions in female cervical mucosa, leveraging advanced techniques such as flow cytometry and single-cell RNA sequencing (scRNA-seq) method. Flow cytometry allows to characterize established subsets of T cells including TRM. Coupled with scRNA seq, a high resolution of molecular data matrix at cell level enables us to explore molecular signatures of unique T cell clusters and individual T cells within the mucosal tissue. The manuscript is under writing, hoping to publish soon.
Postdoctoral Scientific Collaborator (2021-2022)
Swiss Tropical and Public Health Institute Papua New Guinea Institute of Medical Research
I successfully oversaw the National Health Facility Survey 2021, ensuring its timely and effective completion. To further efficient and effective monitoring and reporting, I designed and developed an automated interactive dashboard, providing streamlined and efficient access to ODK-based survey data collection.
As part of the survey, I completed a draft for an automated report, streamlining the reporting process and enhancing efficiency. Recognizing the need for advanced data collection methods, I conceptualized a QR linkage system to be implemented in the upcoming malaria indicator survey. In addition, I contributed technical consultations to both the Institute and the National Technical Working Group for Malaria. We also attempted to assess the accuracy, reliability, and viability of using repeated malaria surveys to calculate under-five mortality as an effective intervention.
Data Viz
Shiny Apps
R Packages
mStats

mStats is a versatile and efficient collection of functions that I have curated and developed to enhance my work in various domains. This collection encompasses a range of statistical tools and utilities that have proven invaluable in my research and analysis endeavors.
As an avid user of mStats, I am delighted to share this valuable resource with the broader community. Together, we can leverage the power of mStats to overcome statistical challenges, drive impactful insights, and advance the frontiers of knowledge in our respective fields.
Stay tuned for updates and enhancements to mStats as I continue to refine and expand its capabilities to meet the evolving needs of the research community.
shinyAuthX

ShinyAuthX is a powerful R package specifically designed for user authentication within Shiny apps. It provides a comprehensive suite of authentication features including user sign-in, sign-up, sign-out, and password recovery, seamlessly integrating user-friendly UI and Server components. It empowers shiny app developers to create secure and user-friendly authentication systems.
u5mr
Introducing u5mr, an open-source R package designed for estimating child mortality rates. This package includes the implementation of various methods to estimate child mortality.
One notable feature of u5mr is its incorporation of the maternal age cohort-derived (MAC) and period-derived (MAP) methods proposed by Rajaratnam et al. These methods provide additional flexibility and accuracy in estimating child mortality rates, enhancing the reliability of the results.
To learn more about the MAC and MAP methods, refer to the publication by Rajaratnam et al. available at Rajaratnam et al.



